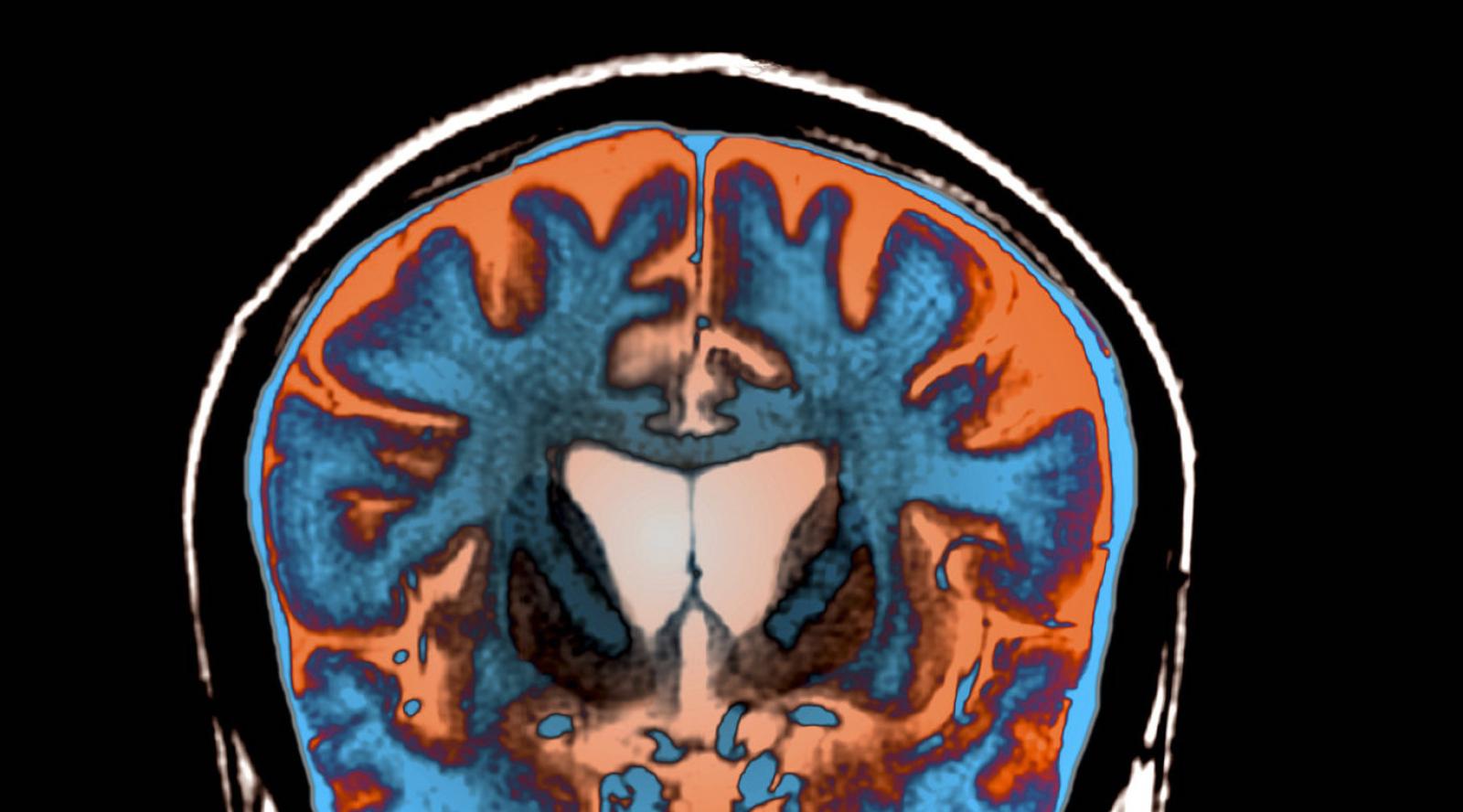
An MRI scan shows signs of atrophy in the brain of a patient with Huntington's disease.
Science Photo Library/Science Source
Researchers have found that aberrant protein aggregates responsible for Huntington’s disease have some weak spots that could be exploited to hinder the development of this pathology. The study, published on Scientific Report, has been conducted by scientists of the Centre for Complexity and Biosystems (CC&B) of the University of Milan, in collaboration with colleagues from Penn State University.
Huntington’s disease is a genetic neurodegenerative disorder caused by the production of an abnormal version of a protein, huntingtin, which is involved in several cellular processes. Mutated huntingtin tends to form aberrant aggregates – like those characteristic of other neurodegenerative disorders – that, as the disease advances, interfere with neuron functions and can also be toxic to certain cell types in the brain.
Recent studies have shown that in Parkinson’s and Alzheimer’s diseases the formation of these lethal protein aggregates can be transmitted to neighbouring cells, with potential prion-like infection and propagation as in the mad cow disease; these aberrant proteins can also induce their normal counterparts to change their structure assuming the aberrant one. Basically, they are capable of “infecting” the normal proteins, thus spreading the mutated conformation. Recently, it has been found that something similar occurs also in the Huntington’s disease but the biological dynamics that leads to the formation of heterogeneous aggregates – i.e. those formed by both normal and mutated huntingtin – have never been studied so far, since most of the research focused on homogeneous aggregates of aberrant huntingtin.
CC&B researchers used their consolidated multidisciplinary approach to study the formation of heterogeneous aggregates of huntingtin, combining computational models and biological experiments. As a first step, they run a particular type of computational simulation that allowed them to investigate protein structures and their possible interactions. In this way, they identified a possible aggregation mechanism through which a single mutated huntingtin can bind to its normal counterparts, and some elements of the protein structure that trigger the aggregation process. After that, they verified their computational results in the laboratory, by adding mutated huntingtin in cell cultures that only possesses normal huntingtin, and analysing them with a special microscopic technique that allowed them to observe the co-localization and likely formation of heterogeneous aggregates of the two protein versions.
«Once we managed to identify the configurations that characterized the aggregates, we could understand which were the most promising molecular targets to destabilize these abnormal protein agglomerations», explained Silvia Bonfanti, post-doctoral researcher at CC&B and first author of the study.
«The identification of these possible “weak spots” in the structure of toxic aggregates of huntingtin could allow the development of drugs capable of hindering their formation, or at least of slowing it down, thus impeding the progress of the disease», added Caterina La Porta, professor of General Pathology at the Department of Environmental Sciences and Policy of the University of Milan and coordinator of the research.